About
Reilly Brennan
Objective
To determine how geographic, genetic, and ecological interactions between rodents in a network can be used to explore the risk of disease spillover.
The System
We are focusing on a rodent-ectoparasite network in the neotropics (Chile), where hantavirus in endemic. There are 50 rodent species documented to be parasitized by a total of 131 different ectoparasites which include fleas, ticks, mites and lice. We updated a review of Chilean rodents and ectoparasites (Landaeta-Aqueveque et al. 2021) to include associations documented until March 2023.
Methods
We use host-ectoparasite associations to build an ecological network of rodent hosts where they are connected by ectoparasites they share. Based on this, we explore the probability for rodents to share ectoparasites based on range overlap, range distance, and phylogenetic relatedness. We then assess how well the network model predicts Hantavirus hosts and use it to assess potential novel hosts.
Maps
- Explore the rodent-ectoparasite network by first creating rodent richness maps for all rodent species in Chile and only rodents that are documented with ectoparasites.
- Create a range overlap and range distance matrix.
Phylogeny
- Assess the phylogenetic relationships between rodent species in the dataset
- Build a phylogeny using Geneious Prime software and export the distance matrix.
Ectoparasite sharing
- Use the GeoTax package (Robles 2023) to calculate the probability for rodents to share ectoparasites based on range overlap, range distance, and phylogenetic relatedness.
- Make plot showing the probability to share ectoparasites for each ectoparasite and for the whole dataset.
Network building
- Use an adjacency matrix to build a network in the igraph package.
- Use the igraph package to find the shortest paths between all pairs of rodents in the network.
Network Analysis
- Test for correlations between network distances with phylogenetic and range distance and between range and phylogenetic distance using mantel tests.
- Use the closeness centrality from Gephi to split the rodents into quantiles and visually plot them based on their groups.
- Test how well closeness centrality predicts hantavirus hosts using a binomial logistic regression.
In addition to the code documented on this site, we used Gephi software (Bastian M., n.d.) to build our rodent-ectoparasite network and to calculate scores of closeness centrality for each rodent. We also used Geneious Prime 2023.0.4 (https://www.geneious.com), to build a neighbor-joining phylogeny based on MUSCLE alignment to caluclate pairwise phylogenetic distance between rodent species. We used cytpchrome b sequences downloaded from GenBank (Sayers et al. 2022).
Network Model
This network shows rodents as nodes (cirles) and shared ectoparasites
as edges (connecting lines). The edges are weighted by how many
ectoparasites the rodents share and the nodes are weighted by the
closeness centrality of the rodent in the network. The colors correspond
to the closeness centrality quantile group that the rodent belongs to
with yellow being the most central and dark blue being the least. We
used Gephi software to construct this model. 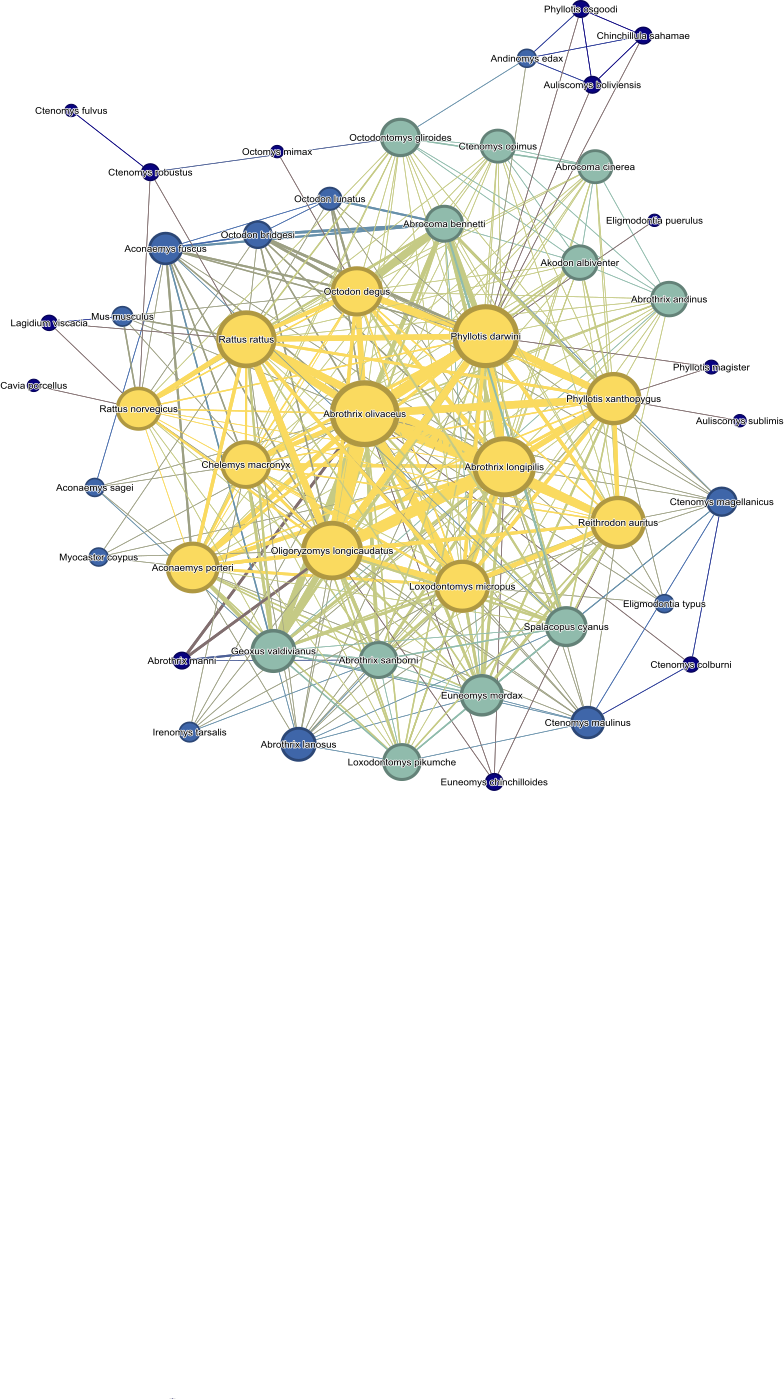
Cited: