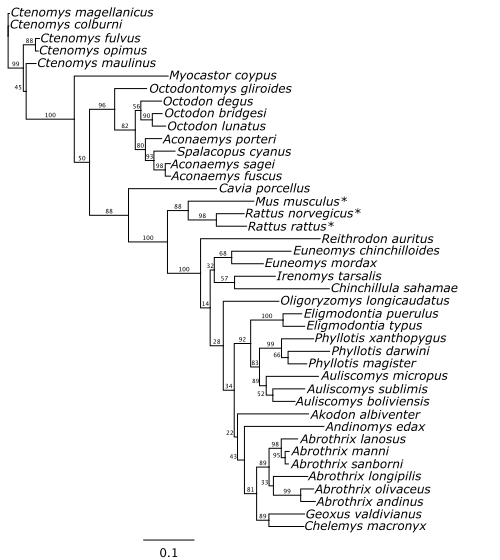
Phylogeny
Purpose: Create a phylogenetic tree of the rodent species to calculate the pairwise genetic distances between rodents.
To calculate pairwise phylogenetic distance for the rodent species in our network, we first downloaded cytochrome b sequences for the rodent species from GenBank (Sayers et al. 2022). We included sequences with at least the first 801 base pairs of cytochrome b (maximum length= 1160 bp) which resulted in sufficient sequences for 42 of the 50 rodent species.Using Geneious Prime 2023.0.4 (https://www.geneious.com), we imported the 42 sequences from GenBank and aligned them using the MUSCLE algorithm. Cutting down to only the first 801 bp of the aligned sequences, we constructed a maximum-likelihood phylogenetic tree with PhyML 3.0 (1000 replications) and obtained patristic distances from the distance matrix option.